
最先端AI技術による高精度/高記述力の潜在空間学習と分子設計
大上 雅史
研究代表者
所属:東京科学大学情報理工学院 准教授
研究概要
本計画研究は、機械学習や化合物設計技術によって天然物から様々な情報を抽出し分子設計を主導するAI技術を開発します。これまでのAI技術では多くの天然物が該当するbRO5 (beyond Rule-of-Five) 空間への対応ができていませんでしたが、bRO5空間への手法の拡充や、ペプチドと合成小分子をつなぐ技術の開発に取り組みます。潜在空間からデザインされた化合物のインシリコスクリーニング評価や、他の計画班による実験的評価を踏まえ、AIによる潜在空間構築をさらに洗練させ、本領域が目指す潜在空間を応用した革新的な生物活性分子設計技術を実現します。
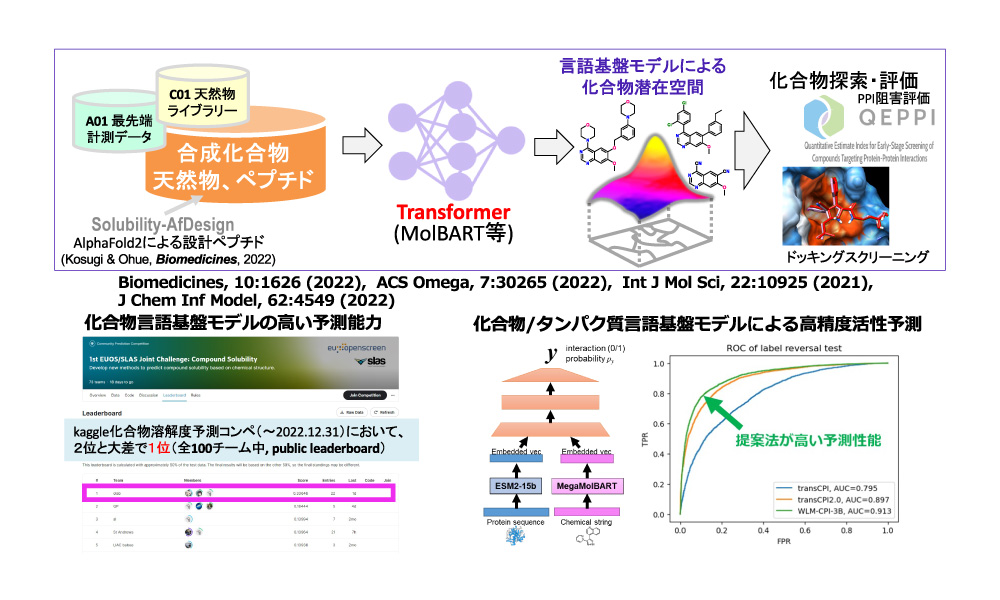
研究成果
- Furui K, Ohue M. ALLM-Ab: Active learning-driven antibody optimization using fine-tuned protein language models. Journal of Chemical Information and Modeling (epub ahead of print). doi: 10.1021/acs.jcim.5c01577
- Kengkanna A, Kikuchi Y, Niwa T, Ohue M. Reaction-conditioned generative model for catalyst design and optimization with CatDRX. Communications Chemistry, 8: 314, 2025. doi: 10.1038/s42004-025-01732-7
- 大上雅史. タンパク質の構造を予測する新技術. ニュートン別冊 新・生命論, 2025.
- Furui K, Ohue M. Boltzina: Efficient and Accurate Virtual Screening via Docking-Guided Binding Prediction with Boltz-2. In Proceedings of AI4Mat Workshop on NeurIPS 2025, 2025. (accepted) doi: 10.48550/arXiv.2508.17555
- Furui K, Sakano K, Ohue M. Predictive and therapeutic applications of protein language models. Allergology International, 74(4): 534-548, 2025. doi: 10.1016/j.alit.2025.08.004
- Igarashi K, Ohue M. Protein–ligand affinity prediction via Jensen–Shannon divergence of molecular dynamics simulation trajectories. Biophysics and Physicobiology, 22(3): e220015, 2025. doi: 10.2142/biophysico.bppb-v22.0015
- Uchikawa K, Furui K, Ohue M. Leveraging AlphaFold2 structural space exploration for generating drug target structures in structure-based virtual screening. Biochemistry and Biophysics Reports, 43: 102110, 2025. doi: 10.1016/j.bbrep.2025.102110
- Yasumitsu Y, Ohue M. Generation of appropriate protein structures for virtual screening using AlphaFold3 predicted protein–ligand complexes. Computational and Structural Biotechnology Reports, 2: 100057, 2025. doi: 10.1016/j.csbr.2025.100057
- Masunaga S, Furui K, Kengkanna A, Ohue M. GraphBioisostere: General Bioisostere Prediction Model with Deep Graph Neural Network. In The 31st Int’l Conf on Parallel and Distributed Processing Techniques and Applications (PDPTA’25).
- Ohue M, Furui K. GBDT-based Model and Web Tool for Prediction of Blood-Placental Barrier Permeability of Small Molecules. In The 31st Int’l Conf on Parallel and Distributed Processing Techniques and Applications (PDPTA’25).
- 大上雅史. タンパク質言語モデルの創薬分野での利用. SAR News, 48: 8-12, 2025.
- Furui K, Ohue M. Benchmarking HelixFold3-Predicted Holo Structures for Relative Free Energy Perturbation Calculations. ACS Omega, 10(11): 11411-11420, 2025. doi: 10.1021/acsomega.4c11413
- Hu W, Ohue M. SpatialPPIv2: Enhancing protein–protein interaction prediction through graph neural networks with protein language models. Computational and Structural Biotechnology Journal, 27: 508-518, 2025. doi: 10.1016/j.csbj.2025.01.022
- 大上雅史. AIが広げる分子設計の可能性. Drug Delivery System, 40(1): 62-70, 2025.
- Furui K, Shimizu T, Akiyama Y, Kimura SR, Terada Y, Ohue M. PairMap: An Intermediate Insertion Approach for Improving the Accuracy of Relative Free Energy Perturbation Calculations for Distant Compound Transformations. Journal of Chemical Information and Modeling, 65(2): 705-721, 2025. doi: 10.1021/acs.jcim.4c01634
- Ohue M, Yasuo N, Takata M. Innovations in Mathematical Modeling, AI, and Optimization Techniques. The Journal of Supercomputing, 81: 340, 2025. doi: 10.1007/s11227-024-06861-9
- Sakano K, Furui K, Ohue M. NPGPT: natural product-like compound generation with GPT-based chemical language models. The Journal of Supercomputing, 81: 352, 2025. doi: 10.1007/s11227-024-06860-w
- 大上雅史. マルチモーダルでマルチモダリティな創薬を支援する情報技術. 実験医学, 43(1): 13-18, 羊土社, 2025. doi: 10.18958/7641-00001-0001766-00
- 大上雅史. AlphaFold2の衝撃. 実験医学別冊 最強のステップUPシリーズ AlphaFold時代の構造バイオインフォマティクス実践ガイド, 15-26, 羊土社, 2024.
- Furui K, Ohue M. Active learning for energy-based antibody optimization and enhanced screening. In Proceedings of Machine Learning in Structural Biology Workshop (MLSB 2024) at NeurIPS 2024, 2024. doi: 10.48550/arXiv.2409.10964
- 大上雅史. AlphaFold3で何ができる? 現代化学 2024年7月号, 東京化学同人, 2024.
- Sakano K, Furui K, Ohue M. Natural Product-like Compound Generation with Chemical Language Models. In Parallel and Distributed Processing Techniques. CSCE 2024. Communications in Computer and Information Science, 2256: 153-166, 2025. doi: 10.1007/978-3-031-85638-9_12
- Kengkanna A, Ohue M. Enhancing property and activity prediction and interpretation using multiple molecular graph representations with MMGX. Communications Chemistry, 7: 74, 2024.
- Furui K, Ohue M. FastLomap: Faster Lead Optimization Mapper Algorithm for Large-Scale Relative Free Energy Perturbation. The Journal of Supercomputing, 80: 14417-14432, 2024.
- Ueki T, Ohue M. Antibody Complementarity-Determining Region Design using AlphaFold2 and DDG Predictor. The Journal of Supercomputing, 80: 11989-12002, 2024.
- Hu W, Ohue M. SpatialPPI: three-dimensional space protein-protein interaction prediction with AlphaFold Multimer. Computational and Structural Biotechnology Journal, 23: 1214-1225, 2024.
- 大上雅史. AlphaFoldによる高精度なタンパク質立体構造予測と創薬への応用. PHARM STAGE, 23(8): 24-28, 2023.
- Murakumo K, Yoshikawa N, Rikimaru K, Nakamura S, Furui K, Suzuki T, Yamasaki H, Nishigaya Y, Takagi Y, Ohue M. LLM Drug Discovery Challenge: A Contest as a Feasibility Study on the Utilization of Large Language Models in Medicinal Chemistry. In Proceedings of AI for Accelerated Materials Design (AI4Mat) NeurIPS 2023 Workshop, 2023.
- Ochiai T, Inukai T, Akiyama M, Furui K, Ohue M, Matsumori N, Inuki S, Uesugi M, Sunazuka T, Kikuchi K, Kakeya H, Sakakibara Y. Variational autoencoder-based chemical latent space for large molecular structures with 3D complexity. Communications Chemistry, 6: 249, 2023.
- Kosugi T, Ohue M. Design of cyclic peptides targeting protein–protein interactions using AlphaFold. International Journal of Molecular Sciences, 24(17): 13257, 2023.
- Ohue M, Kojima Y, Kosugi T. Generating potential protein-protein interaction inhibitor molecules based on physicochemical properties. Molecules, 28(15): 5652, 2023.
- Kengkanna A, Ohue M. Enhancing Model Learning and Interpretation Using Multiple Molecular Graph Representations for Compound Property and Activity Prediction. In Proceedings of The 20th IEEE International Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB 2023), 2023.
- Furui K, Ohue M. Faster Lead Optimization Mapper Algorithm for Large-Scale Relative Free Energy Perturbation. In Proceedings of The 29th International Conference on Parallel & Distributed Processing Techniques and Applications (PDPTA’23), 2023.
- Kosugi T, Ohue M. Solubility-aware protein binding peptide design using AlphaFold. Biomedicines, 10(7): 1626, 2022.
- Kosugi T, Ohue M. Quantitative estimate index for early-stage screening of compounds targeting protein-protein interactions. International Journal of Molecular Sciences, 22(20): 10925, 2021.
